
Early Evolution of Translation - The Reverse Recognition Conjecture
1. Hypothesis on the origin of the genetic code:
Our contemporary
triplet genetic code was preceded by an
ancient doublet code
(Wilhelm
T and Nikolajewa S, 2004,
Patel 2005,
Wu et al, 2005 ,
Copley et al, 2005 ).
However, in
order to avoid the frameshift problem one has to assume a triplet reading
frame also in doublet coding times.
One still wonders about
such information wasting, where the third base would not carry any
information at all.
2. Hypothesis on the early evolution of translation: early pre-tRNA (or tRNA precursor) binding was independent of the 5'-3' direction:
Figure 1. Only two bases bound,
either at the first two codon nucleotides, or at the last two ones. In this
scenario all pre-mRNA nucleotides carry information, needed to translate the
codon.

Figure 2. Two different tRNAs could recognize the same codon
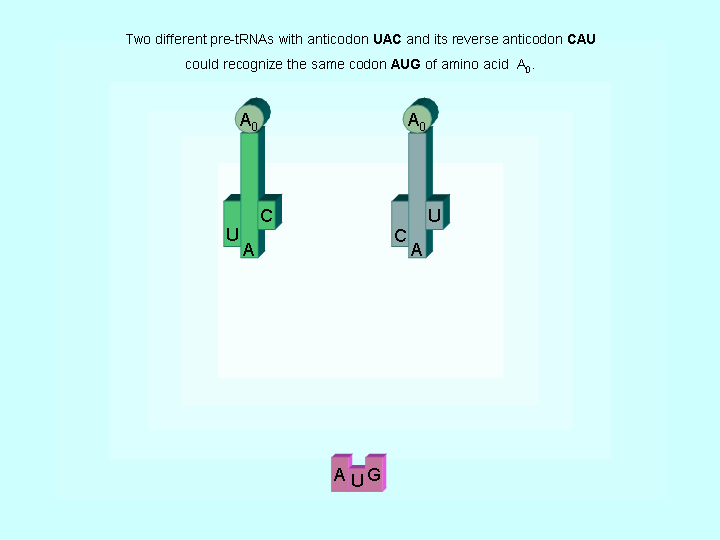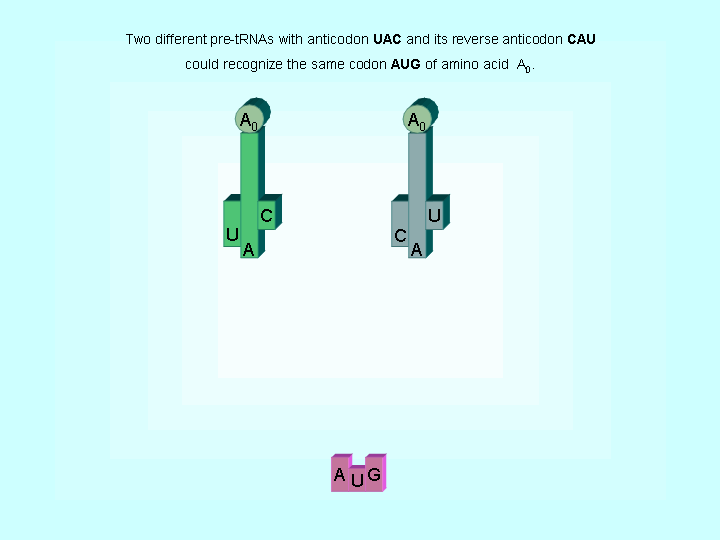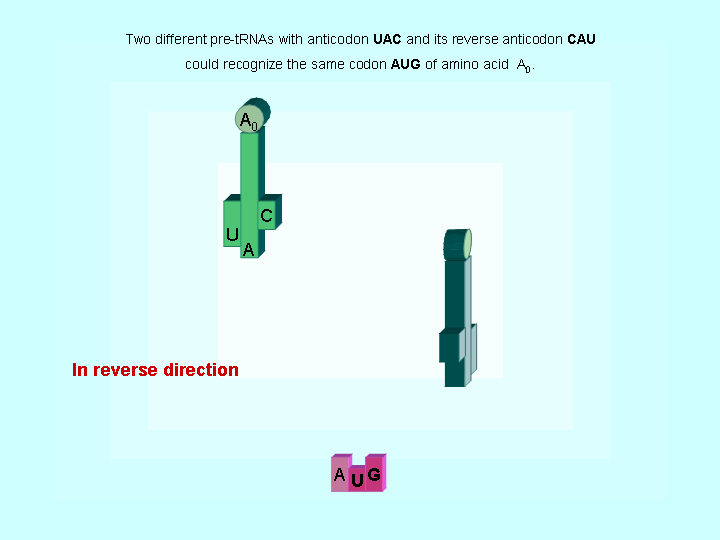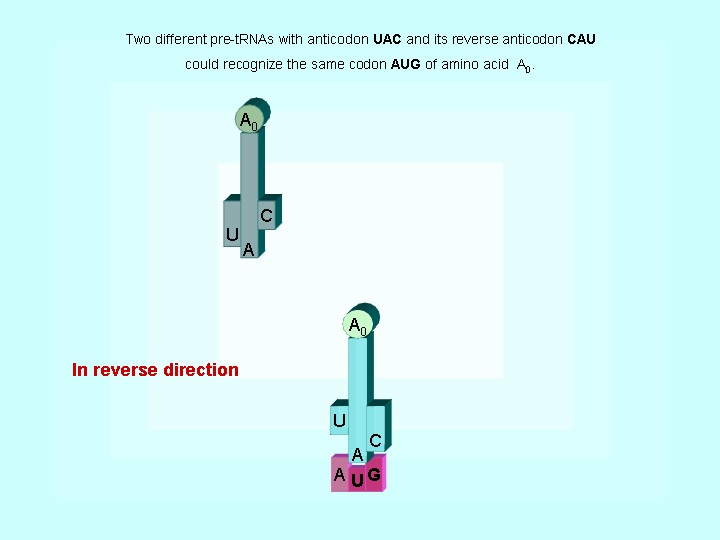
| The evidences to support the hypothesis of early translations | |
| Order in the Genetic Code | |
| Amino acids that have similar biochemical properties tend to have similar codons (The new classification scheme of the genetic code and codon symmetry of the genetic code) | |
| Special role of the second base of codons | |
|
|
| Evolutionary conserved group of amino acids | |
| From analysis of the amino-acid
replacement matrices, observation that reverse codon pairs (XYZ and ZYX
codons) generally encode
evolutionary similar amino acids (Thompson
et al,1994) . We suppose that this
observation is a relict from old ``reverse recognition times'', where the
reverse recognition should have a minimal effect on the resulting
polypeptide. Strongly conserved groups of amino acids (Thompson et al, 1994) are subsets of exactly one quadrant, e.g. the amino acids M, I, L, V belong to the upper right block in the new classification table of the genetic code. The other conserved strong groups belonging to one block of the new scheme are MILF, STA, NEQK, NHQK, NDEQ, HY. The only exceptions are QHRK (R (Arg) is in another quadrant) and FYW (in three quadrants). In other words, reverse codon pairs tend to code for evolutionary similar amino acids, and each quadrant is enriched for amino acids with similar biochemical properties. |
|
| Observation about STOP codons | |
| Reverse STOP codons (GAT, AGT, AAT) do not have their own tRNAs (just one exception in human). | |
| Observation about tRNA | |
| Ancient pre-tRNAs presumably only consisted of the anticodon loop, lacking the D- and T-loops (Rodin et al, 1993). Such pre-tRNAs would have been (almost) symmetrical and could thus bind in two directions. If the reverse recognition model is correct, the resulting polypeptide should be relatively independent of the pre-tRNA binding direction. | |
| Observation about tRNA anticodons prevalence | |
|
1) Of course, no species has a tRNA with an anticodon complementary to any termination codon. Intriguingly, there is also no tRNA with an anticodon for a reverse STOP codon (GAT, AGT, AAT). The only exception is H. sapiens with one tRNAAsn with the anticodon ATT. The lack of specific tRNAs does not imply that no tRNA exists which can recognize reverse STOP codons. For instance, using base pairing allowed by Crick's wobble rules, tRNA with the GTT anticodon can recognize the reverse STOP codon AAT. 2). The significant suppression of tRNAs with A at the first anticodon position. A** anticodons are fully excluded in archaea. In bacteria and eucaryotes there are some exceptions, but it can be observed that AY* anticodons do not appear in any species. 3). The significant suppression of A*A self-reverse codons. In no archaea and in no bacteria any tRNA has such an anticodon. In archaea the anticodon TAT is the only one without own tRNAs that is not a STOP anticodon or an A** anticodon. Interestingly, this is the only anticodon which according to Crick's wobble rules allows recognition of a STOP codon (TAG)
|
|